SPSED Signal Peptide Secretion Efficiency Database
About Signal Peptide
Signal peptides (SPs) are short peptides directing newly synthesized proteins towards the secretory pathway. SPs are usually 16~30 amino acids long and are found in the N-terminus of proteins in virtually all organisms. Signal peptidases will remove SPs after the protein translocation.
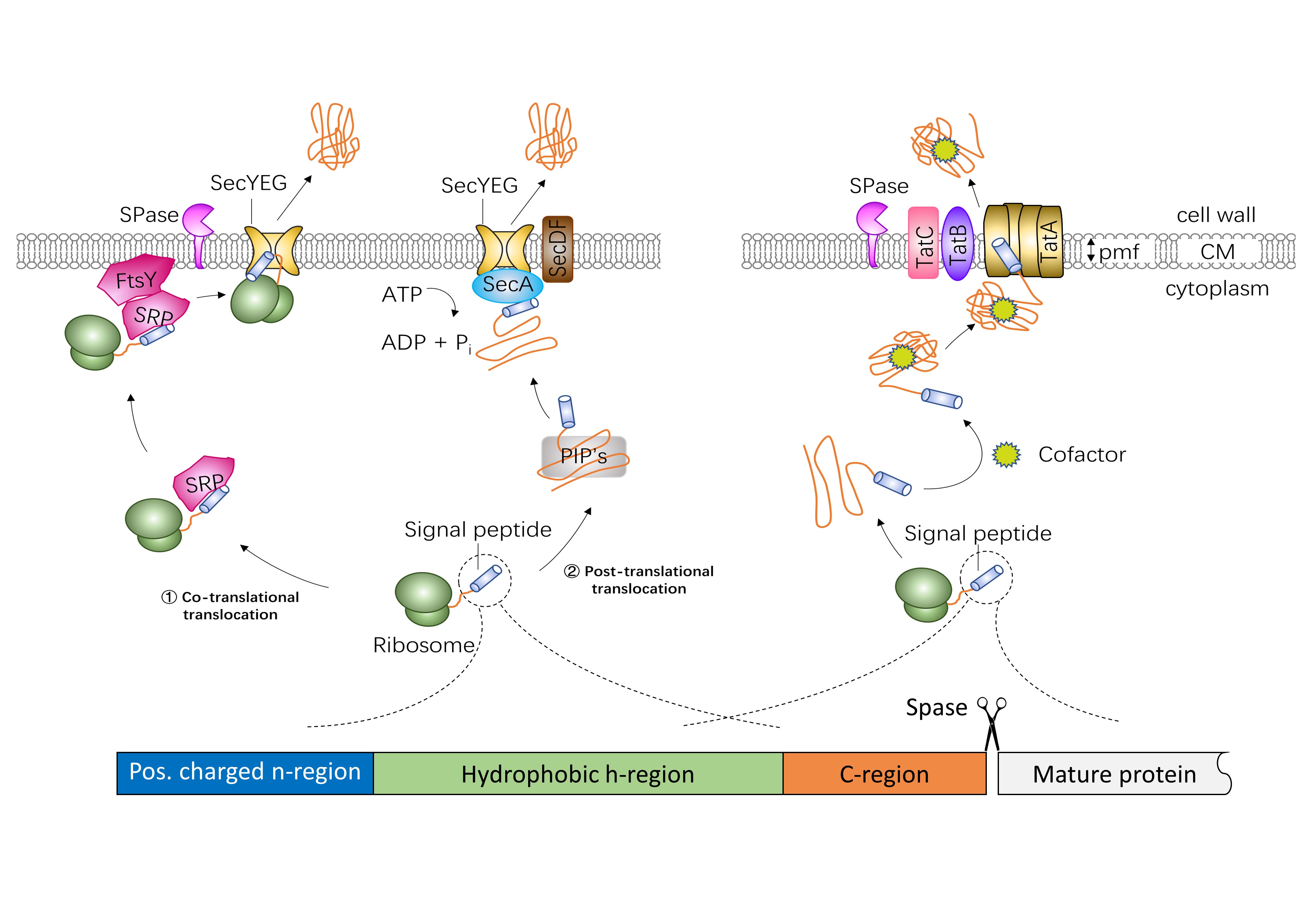
Several different protein export systems exist in bacteria. The general secretion (Sec) protein export pathway and the twin-arginine translocation (Tat) export pathway are the two main protein secretion pathways. Sec-dependent proteins are translocated to the plasma membrane either co- or post-translationally. In the co-translational export mode, precursor proteins are recognized at the ribosome by the signal recognition particle (SRP) and then targeted to the transmembrane SecYEG channel by SRP and FtsY, the SRP membrane receptor. In the post-translational export mode, the post-translationally interacting proteins (PIP’s), such as the specific targeting chaperone SecB, the general chaperones GroELS, DnaK-DnaJ-GrpE, trigger factor, the CsaA protein and the soluble form of SecA, keep the fully synthesized precursor proteins in an unfolded secretion-competent state. Then the motor protein SecA translocate the preproteins through SecYEG using metabolic energy from ATP hydrolysis. In addition, SecDF enhances the release of preproteins. Tat-dependent proteins are transported across lipid bilayers in a folded state. The energy for translocation comes from the proton motive force (PMF). In gram-negative bacteria and gram-positive bacteria with high GC-content genomes, the Tat translocase consists of TatA, TatB, and TatC. In low-GC gram-positive bacteria, the Tat system is composed of TatC and a bifunctional TatA protein.
Signal peptides from different proteins show a common structure. Generally, a signal peptide is composed of three distinct domains: a positively charged n-region (1-5 residues long), a central, hydrophobic h-region (7-15 residues long) and a c-region (3-7 residues) with the cleavage site of signal peptidase. A highly conserved twin-arginine motif (SRRXFLK, where X is often, but not always, a polar amino acid residue) is located at the n/h-region boundary of Tat-specific signal peptides.
Introduction for SPSED
As an important secretory functional element of bacterial cell factories, the adaptation of signal peptide to heterologous protein will significantly affect the secretion level of heterologous protein. In recombinant protein production, screening an optimum signal peptide is an important but toilsome step to increase the yield of the target protein. However, it's currently not possible to predict the best-performing signal peptide for a given target protein. The non-integrated and fragmented data in relevant literature hindered the systematic investigation of protein secretion efficiency guided by the signal peptide.
SPSED is a database of signal peptide secretion efficiency for specific target proteins. One entry in the database corresponds to the secretion yield of a specific target protein with a specific signal peptide. We hope that SPSED is helpful in the production of enzymes and the research of signal peptide secretion mechanism.
What you can do with SPSED
Citation
Peng C, Guo Y, Ren S, Li C, Liu F and Lu F (2022) SPSED: A Signal Peptide Secretion Efficiency Database. Front. Bioeng. Biotechnol. 9:819789. doi: 10.3389/fbioe.2021.819789. [Link]
Feedback
-
1.0
Jun.28,2020
SPSED 1.0
The download page was added; more data was added.
-
0.8
May.18,2019
SPSED 0.8
The pages were re-designed and the layout of home page was modified. The submit page was added.
-
0.5
April.23,2019
SPSED 0.6
The backend function test of the website completed locally and the online test started. SPSED can be visited through www.spsed.com.
-
0.1
Jan.09,2019
SPSED 0.1
The data was collected and the initial construction of the website began.