SPSED Signal Peptide Secretion Efficiency Database
This page gives clear instructions on how to use SPSED. If you have any problems, please do not hesitate to contact Dr. Chong Peng. Email: cpeng@tust.edu.cn
Home Page
The home page provides a basic introduction to signal peptide and SPSED (1). Users can fill the target protein or expression host in the search box to make a quick search in the database (2). This page also presents an overview of the dataset in the SPSED database (3) and some commonly used links about signal peptides (4).

Browse page
The browse page of the database preferentially displays secreted protein, the number of signal peptides fused to this secreted protein, the expression host, and the related references for each record (1). Upon clicking any secreted protein, the signal peptide sequence, signal peptide type, and the corresponding target protein yield is displayed in another browser page (2). In the new browser page, users can click a SPSED ID to obtain the detailed information of the entry (4). The nucleotide sequence, amino acid sequence, Uniprot link, GenBank link, and expression host of the secreted target protein have been displayed in both ‘Secreted Protein Detail’ page (3) and ‘Detail Information’ page (4) for each entry. As for the signal peptide fused to the protein, the signal peptide source, type, sequence, DNA, and protein sequence of signal peptide original protein have been provided. We also calculated the hydrophobicity of the whole signal peptide and the hydrophobicity of h-region with values according to the Kyte-Doolittle hydrophobic scale. The hydrophobicity plot of the whole signal peptide sequence is given on this page. The charge of the whole signal peptide and charge of n-region is also calculated and provided on the page. We used the ratio of current yield to the highest yield to represent the secretion capacity of each signal peptide (4).
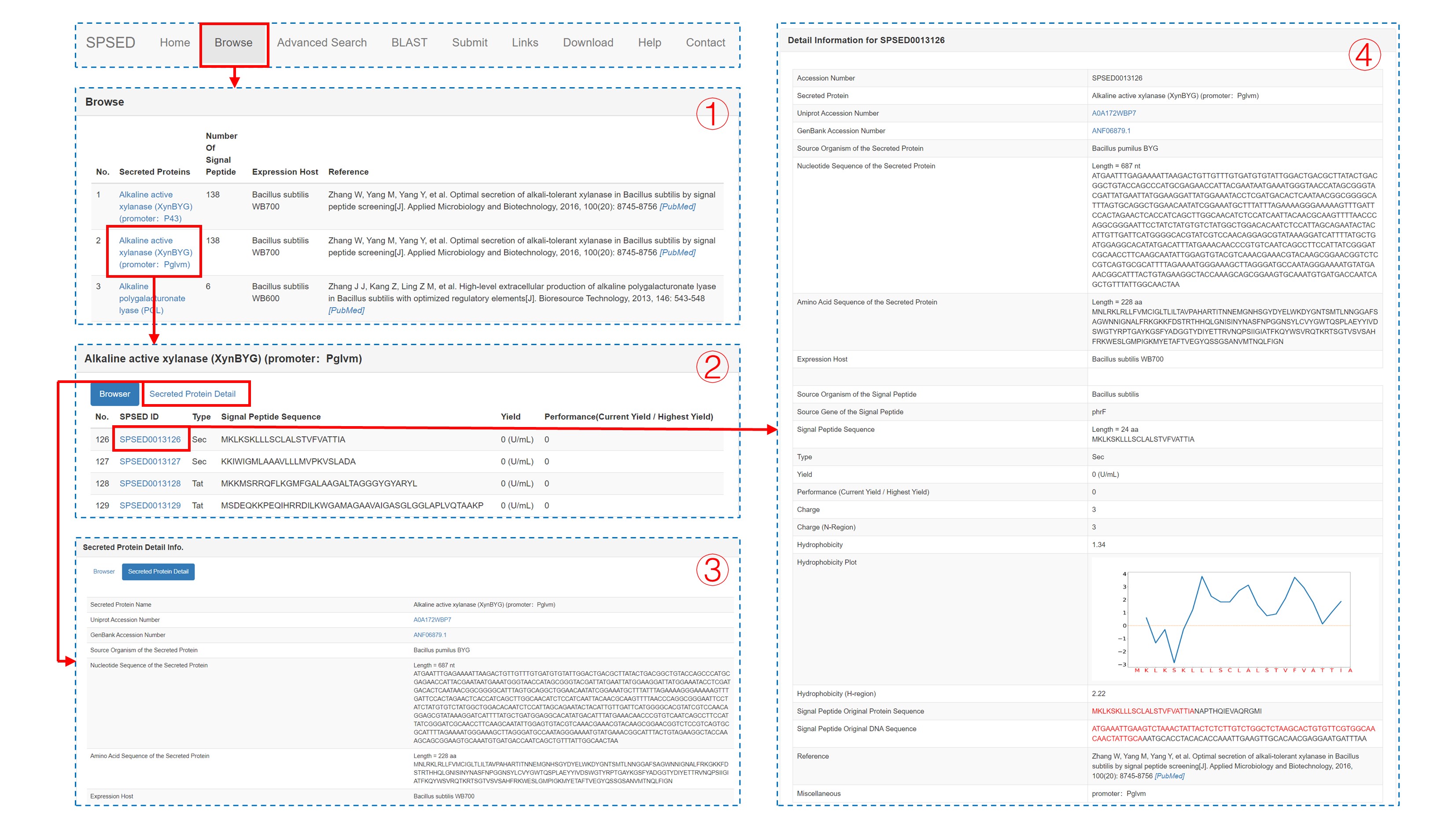
Advanced Search page
This page is designed for a customizable search of SPSED records. Users can select the options listed in the five select boxes named ‘Secreted Protein’, ‘Expression Host’, ‘Signal Peptide Type’, ‘Source Organism of the Signal Peptide’, and ‘Screening of Secretory Efficiency’ to filter out the records they are interested in (1). Upon clicking the SPSED ID listed in the Advanced Search Result page (2), the signal peptide detail information page (3) will be displayed.
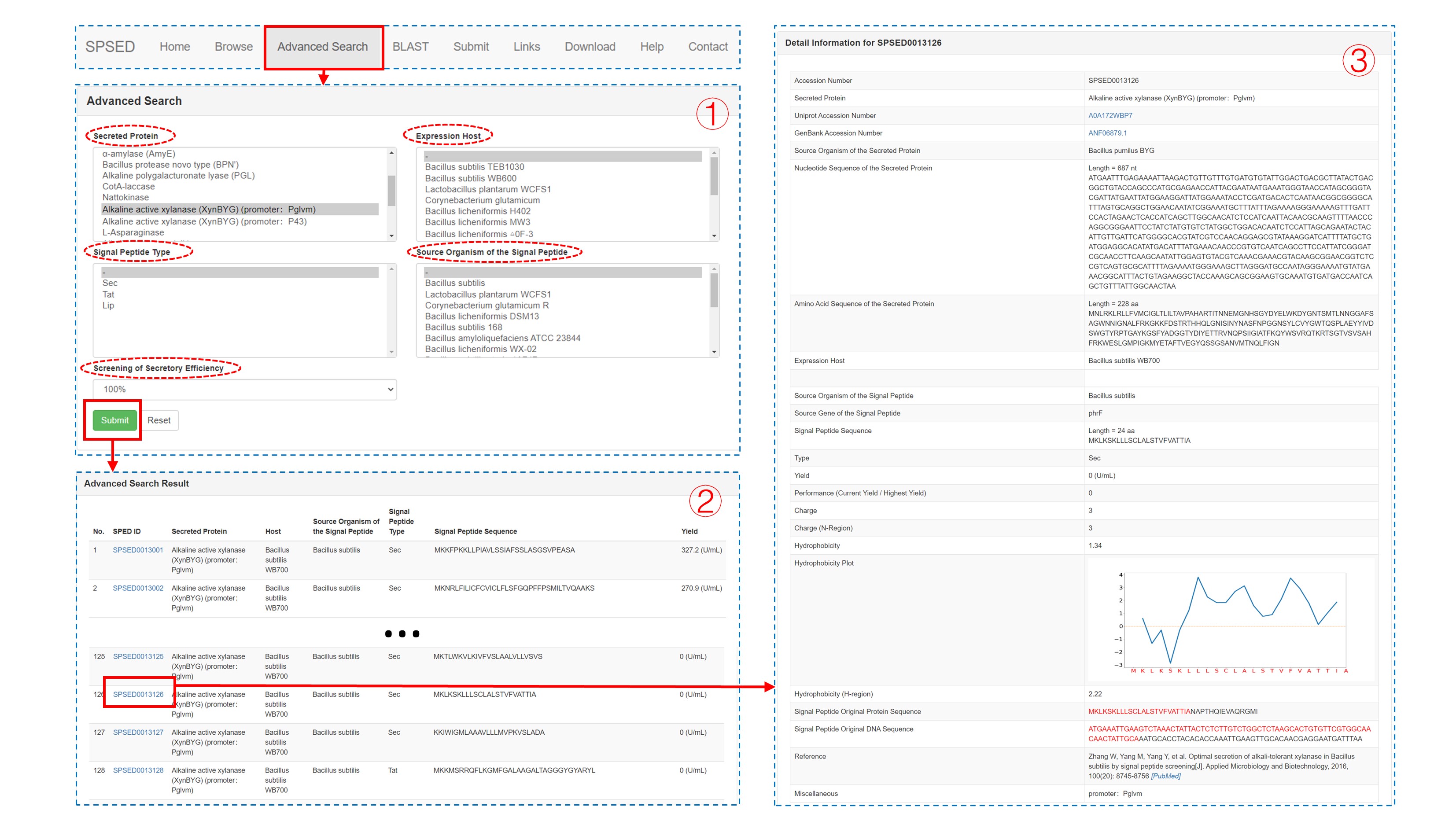
BLAST page
We have installed the BLAST program locally. When the protein or signal peptide of interest is not in SPSED, users can BLAST the query sequence against our database to find a homologous secreted protein or signal peptide. The query amino acid sequence is required to be pasted in the textbox in fasta format (1). When the alignment is ready, a BLAST result page with links to the database entries is provided (2). Upon clicking the SPSED ID listed in the BLAST result page, users can reach the ‘Secreted Protein Detail’ page (3) and further reach the ‘Detail Information’ page (4).
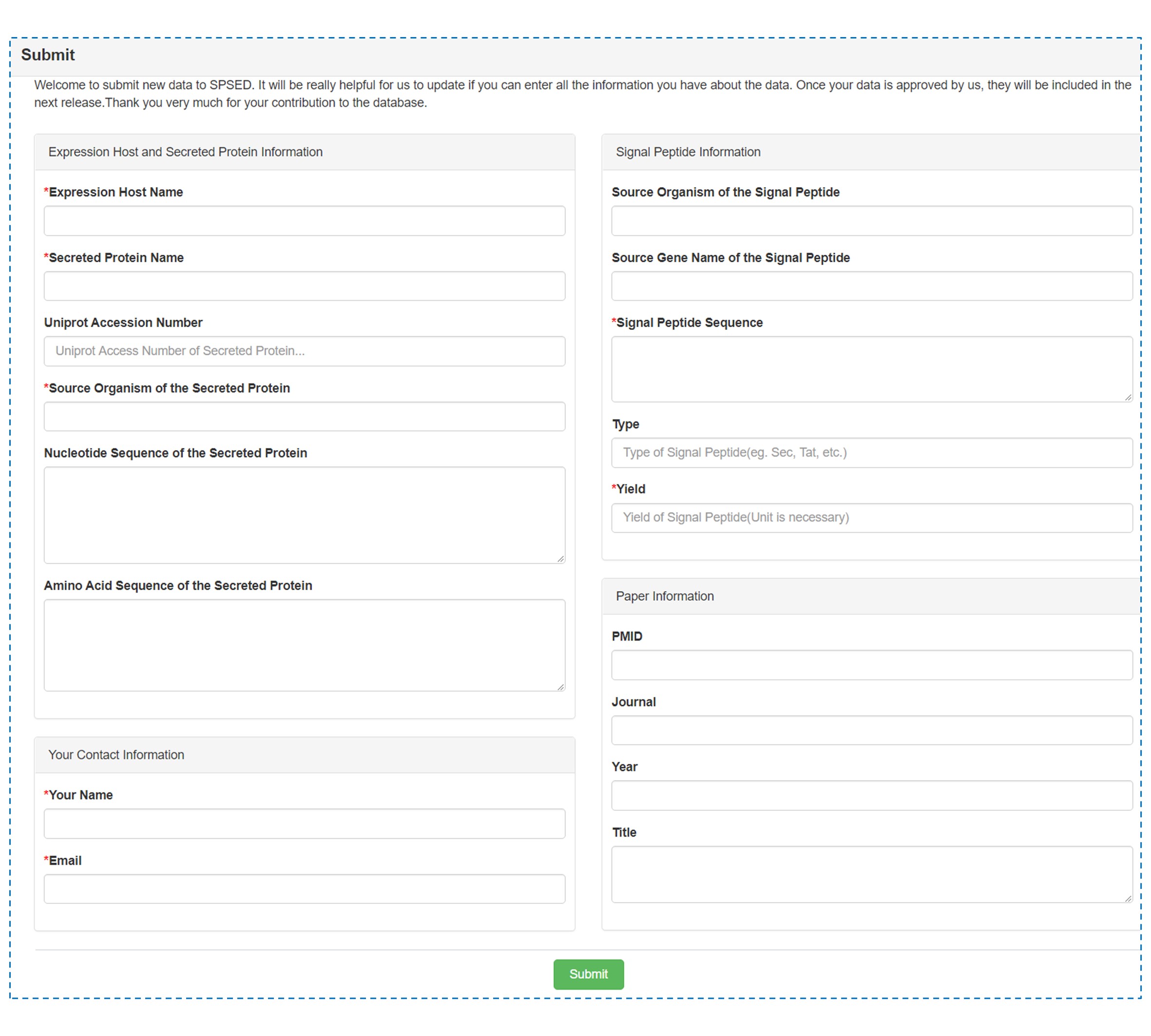
Submit page
Welcome to submit new data to the SPSED database. Please enter all the information you have about the data to help us add a new record. The submitted data will be manually revised and incorporated into the release of the SPSED database. Thank you for your contribution.
Links page
This page provides a list of tools for signal peptide prediction and web resources related to protein secretion.
Download page
Data about target proteins yield that is driven by different signal peptides are packaged by the expression hosts and the target proteins. The compressed data are presented on the 'Download' page for batch downloading.